Dacomitinib is a highly selective, orally bioavailable small-molecule inhibitor of the HER family of tyrosine kinases with potential antineoplastic activity. Dacomitinib specifically and irreversibly binds to and inhibits human Her-1, Her-2, and Her-4, resulting in the proliferation inhibition and apoptosis of tumor cells that overexpress these receptors.
Dacomitinib Anhydrous C24H25CLFN5O2 Structure Essay
Dacomitinib Anhydrous C24H25CLFN5O2 Structure Essay
 C24H25ClFN5O2 structure
C24H25ClFN5O2 structure| Molecular Formula | C24H25ClFN5O2 |
| Average mass | 469.939 Da |
| Density | 1.3±0.1 g/cm3 |
| Boiling Point | 665.7±55.0 °C at 760 mmHg |
| Flash Point | 356.4±31.5 °C |
| Molar Refractivity | 129.5±0.3 cm3 |
| Polarizability | 51.3±0.5 10-24cm3 |
| Surface Tension | 62.2±3.0 dyne/cm |
| Molar Volume | 349.5±3.0 cm3 |
Dacomitinib
| PubChem CID: | 11511120 |
|---|---|
| Structure: |
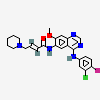  Find Similar Structures
|
| Molecular Formula: | C24H25ClFN5O2 |
| Synonyms: |
Dacomitinib 1110813-31-4 PF299804 Dacomitinib (PF299804, PF299) PF-00299804 More…
|
| Molecular Weight: | 469.9 g/mol |
| Dates: |
|
|
Dacomitinib is a multi-kinase receptor inhibitor used in the therapy of cases of non-small cell lung cancer that harbor activating mutations in the epidermal growth factor receptor gene (EGFR). Dacomitinib is associated with high rate of transient serum aminotransferase elevations during therapy but has not been linked to instances of clinically apparent acute liver injury. Dacomitinib is a member of the class of quinazolines that is 7-methoxyquinazoline-4,6-diamine in which the amino group at position 4 is substituted by a 3-chloro-4-fluorophenyl group and the amino group at position 6 is substituted by an (E)-4-(piperidin-1-yl)but-2-enoyl group. It has a role as an epidermal growth factor receptor antagonist and an antineoplastic agent. It is a member of quinazolines, a member of piperidines, an enamide, a member of monochlorobenzenes, a member of monofluorobenzenes, a tertiary amino compound, a secondary amino compound and a secondary carboxamide.Dacomitinib Anhydrous C24H25CLFN5O2 Structure Essay |
|
1Structures
New Window
1.12D Structure
New Window
|
Chemical Structure Depiction
|
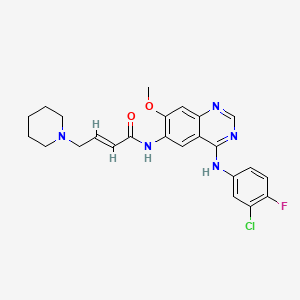 |
|---|
1.23D Conformer
New Window
|
Interactive Chemical Structure Model
|
|
|---|
1.3Crystal Structures
New Window
| PDBe Ligand Code | 1C9 | ||
|---|---|---|---|
| PDBe Structure Code | 4I23 | ||
| PDBe Conformer |
|
2Names and Identifiers
New Window
2.1Computed Descriptors
New Window
2.1.1IUPAC Name
New Window
(E)-N-[4-(3-chloro-4-fluoroanilino)-7-methoxyquinazolin-6-yl]-4-piperidin-1-ylbut-2-enamide
2.1.2InChI
New Window
InChI=1S/C24H25ClFN5O2/c1-33-22-14-20-17(24(28-15-27-20)29-16-7-8-19(26)18(25)12-16)13-21(22)30-23(32)6-5-11-31-9-3-2-4-10-31/h5-8,12-15H,2-4,9-11H2,1H3,(H,30,32)(H,27,28,29)/b6-5+
2.1.3InChI Key
New Window
LVXJQMNHJWSHET-AATRIKPKSA-N
2.1.4Canonical SMILES
New Window
COC1=C(C=C2C(=C1)N=CN=C2NC3=CC(=C(C=C3)F)Cl)NC(=O)C=CCN4CCCCC4
2.1.5Isomeric SMILES
New Window
COC1=C(C=C2C(=C1)N=CN=C2NC3=CC(=C(C=C3)F)Cl)NC(=O)/C=C/CN4CCCCC4
2.2Molecular Formula
New Window
C24H25ClFN5O2
2.3Other Identifiers
New Window
2.3.1CAS
New Window
1110813-31-4
2.3.2Deprecated CAS
New Window
1006885-28-4
2.3.3UNII
New Window
2XJX250C20
2.3.4DSSTox Substance ID
New Window
DTXSID50149493
2.3.5Wikipedia
New Window
Dacomitinib
2.4Synonyms
New Window
2.4.1MeSH Entry Terms
New Window
dacomitinib
N-(4-(3-chloro-4-fluoroanilino)-7-methoxy-6-quinazolinyl)-4-(1-piperidinyl)-2-butenamide
PF 00299804
PF-00299804
PF00299804
Vizimpro
2.4.2Depositor-Supplied Synonyms
New Window
Dacomitinib
1110813-31-4
PF299804
Dacomitinib (PF299804, PF299)
PF-00299804
UNII-2XJX250C20
pf00299804
(2E)-N-[4-[(3-Chloro-4-fluorophenyl)amino]-7-methoxy-6-quinazolinyl]-4-(1-piperidinyl)-2-butenamide
(E)-N-(4-((3-chloro-4-fluorophenyl)aMino)-7-Methoxyquinazolin-6-yl)-4-(piperidin-1-yl)but-2-enaMide. Dacomitinib Anhydrous C24H25CLFN5O2 Structure Essay .
2XJX250C20
PF 00299804-03
PF-00299804-03
C24H25ClFN5O2
(2e)-N-{4-[(3-Chloro-4-Fluorophenyl)amino]-7-Methoxyquinazolin-6-Yl}-4-(Piperidin-1-Yl)but-2-Enamide
J-500784
Dacomitinib [USAN:INN]
Vizimpro
1042385-75-0
dacomitinibum
DACOMITINIB HYDRATE
PF299
(E)-N-[4-[(3-Chloro-4-fluorophenyl)amino]-7-methoxyquinazolin-6-yl]-4-(piperidin-1-yl)but-2-enamide
Dacomitinib (INN)
Dacomitinib anhydrous
PF-299804
PF 00299804
cc-194
Dacomitinib (PF299804)
MLS006011275
GTPL7422
CHEMBL2110732
PF-00299804 dacomitinib
CHEBI:91466
DTXSID50149493
EX-A030
QCR-174
CHEBI:132268
BDBM112499
AOB87383
AOB87735
BCP02530
ABP000126
PF-299
s2727
ZINC72266312
AKOS025401818
CCG-264987
CS-0500
DB11963
SB21754
US8623883, No. 2
NCGC00263185-09
AC-25915
AS-57686
HY-13272
SC-94589
SMR004703025
PF299804|||PF299
AB0035896
D5450
SW219155-1
Y0338
D09883
Dacomitinib (PF299804, PF-00299804)
PF-299804 (Dacomitinib PF-00299804)
Q-4059
Q17130597
(2E)-N-[4-(3-chloro-4-fluoroanilino)-7-methoxyquinazolin-6-yl]-4-(piperidin-1-yl)but-2-enamide
(E)-N-[4-[(3-chloro-4-fluorophenyl)amino]-7-methoxyquinazolin-6-yl]-4-piperidin-1-ylbut-2-enamide
2-Butenamide, N-(4-((3-chloro-4-fluorophenyl)amino)-7-methoxy-6-quinazolinyl)-4-(1-piperidinyl)-, (2E)-
3Chemical and Physical Properties
New Window
3.1Computed Properties
New Window
| Property Name | Property Value | Reference |
|---|---|---|
| Molecular Weight | 469.9 g/mol | Computed by PubChem 2.1 (PubChem release 2019.06.18) |
| XLogP3-AA | 4.4 | Computed by XLogP3 3.0 (PubChem release 2019.06.18) |
| Hydrogen Bond Donor Count | 2 | Computed by Cactvs 3.4.6.11 (PubChem release 2019.06.18) |
| Hydrogen Bond Acceptor Count | 7 | Computed by Cactvs 3.4.6.11 (PubChem release 2019.06.18) |
| Rotatable Bond Count | 7 | Computed by Cactvs 3.4.6.11 (PubChem release 2019.06.18) |
| Exact Mass | 469.168081 g/mol | Computed by PubChem 2.1 (PubChem release 2019.06.18) |
| Monoisotopic Mass | 469.168081 g/mol | Computed by PubChem 2.1 (PubChem release 2019.06.18) |
| Topological Polar Surface Area | 79.4 Ų | Computed by Cactvs 3.4.6.11 (PubChem release 2019.06.18) |
| Heavy Atom Count | 33 | Computed by PubChem |
| Formal Charge | 0 | Computed by PubChem |
| Complexity | 665 | Computed by Cactvs 3.4.6.11 (PubChem release 2019.06.18) |
| Isotope Atom Count | 0 | Computed by PubChem |
| Defined Atom Stereocenter Count | 0 | Computed by PubChem |
| Undefined Atom Stereocenter Count | 0 | Computed by PubChem |
| Defined Bond Stereocenter Count | 1 | Computed by PubChem |
| Undefined Bond Stereocenter Count | 0 | Computed by PubChem |
| Covalently-Bonded Unit Count | 1 | Computed by PubChem |
| Compound Is Canonicalized | Yes | Computed by PubChem (release 2019.01.04) |
3.2Experimental Properties
New Window
3.2.1Boiling Point
New Window
665.7 ºC at 760 mmHg
3.2.2Melting Point
New Window
184-187 ºC
3.2.3Solubility
New Window
<1 mg/mL
3.2.4Octanol/Water Partition Coefficient
New Window
3.92
4Related Records
New Window
4.1Related Compounds with Annotation
New Window
4.2Related Compounds
New Window
| Same Connectivity | 6 Records |
|---|---|
| Same Stereo | 4 Records |
| Same Isotope | 3 Records |
| Same Parent, Connectivity | 9 Records |
| Same Parent, Stereo | 6 Records |
| Same Parent, Isotope | 5 Records |
| Same Parent, Exact | 2 Records |
| Mixtures, Components, and Neutralized Forms | 2 Records |
| Similar Compounds | 2,925 Records |
| Similar Conformers | 191 Records |
4.3Substances
New Window
4.3.1Related Substances
New Window
| All | 122 Records |
|---|---|
| Same | 104 Records |
| Mixture | 18 Records |
Dacomitinib Anhydrous C24H25CLFN5O2 Structure Essay
