Mitochondrial Localization Of Viral Proteins Essay
Mitochondrial Localization Of Viral Proteins Essay
Question:
Discuss About The Mitochondrial Localization Of Viral Proteins.
Answer:
Introduction
The following is a summary of a peer review journal article. Piller et al.,(1999) compile a study titled, “The Amino-Terminal Region for Vpr from Human Immunodeficiency Virus Type 1 Forms Ion Channels and Kills Neurons.” The study is a qualitative kind of research that uses mixed methods research to gather findings. Piller et al.,(1999) seek to examine the terminal for the amino regions for management originating from human HIV. Mitochondrial Localization Of Viral Proteins Essay.he study employs the use of mutagenesis directed to the site of infection together with synthetic peptides. Piller et al.,(1999) establishe various substantial findings. The study identifies the structural regions which are charged with the above metabolic functions.
The study posits that there have been previous attempts to report on the accessory Vpr protein from the HIV. Piller et al.,(1999) find that the channels of activity are altered by the changes due to mutations in the region of the N-terminal of the Vpr (Piller et al., 1999). The variations that occur in the hydrophobic region of the Vpr or the amino acids that range between 53 and 71 do not affect the activity of the N-terminal. Piller et al.,(1999) use the initials to mean Human resource management Immunodeficiency Virus responsible for AIDS infection (Castanier & Arnoult, 2011). The HIV type 1 virus forms channels of action-selection in planar bilayers of lipids. The virus can depolarize neurons that are intact and depolarized. It causes an inward sodium current that results in the death of cells (Castanier & Arnoult, 2011).
Piller et al.,(1999) analyzed the mutations that contain changes in the primary C terminus. The results confirmed previous findings that suggested the region was responsible for the rectification that was observed in the wild versions of the Vpr currents. Piller et al.,(1999) find that a peptide that comprises the first 40 N-terminal amino acids of Vpr is enough for the formation of a channel of ions that are the same as ions that result from the wild type of Vpr found in the planar layers of lipids. Mitochondrial Localization Of Viral Proteins Essay.
ORDER A PLAGIARISM-FREE PAPER NOW
References
Castanier, C. & Arnoult, D., 2011. Mitochondrial localization of viral proteins as a means to subvert
Viruses have developed a battery of distinct strategies to overcome the very sophisticated defense mechanisms of the infected host. Throughout the process of pathogen–host co-evolution, viruses have therefore acquired the capability to prevent host cell apoptosis because elimination of infected cells via apoptosis is one of the most ancestral defense mechanism against infection. Conversely, induction of apoptosis may favor viral dissemination as a result of the dismantlement of the infected cells. Mitochondria have been long recognized for their key role in the modulation of apoptosis but more recently, mitochondria have been shown to serve as a crucial platform for innate immune signaling as illustrated by the identification of MAVS. Thus, it is therefore not surprising that this organelle represents a recurrent target for viruses, aiming to manipulate the fate of the infected host cell or to inhibit innate immune response. In this review, we highlight the viral proteins that are specifically targeted to the mitochondria to subvert host defense. This article is part of a Special Issue entitled Mitochondria: the deadly organelle.
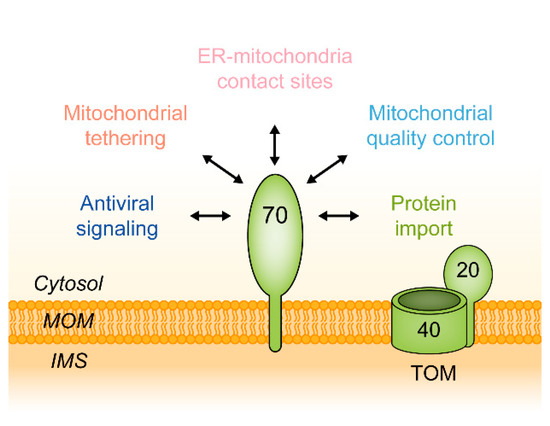
2. Molecular Structure of Tom70
2.1. The Structure of Monomeric Tom70

2.2. Putative Preprotein Binding Sites
2.3. The Oligomeric State of Tom70
3. Evolution of Tom70
3.1. Tom70 and Its Homologs

3.2. The TPR Domain of Tom70 and Its Functional Analogs in Other Membranes
